| Home | Instructions | Output format | Data | Software |
What is LOCALIZER?
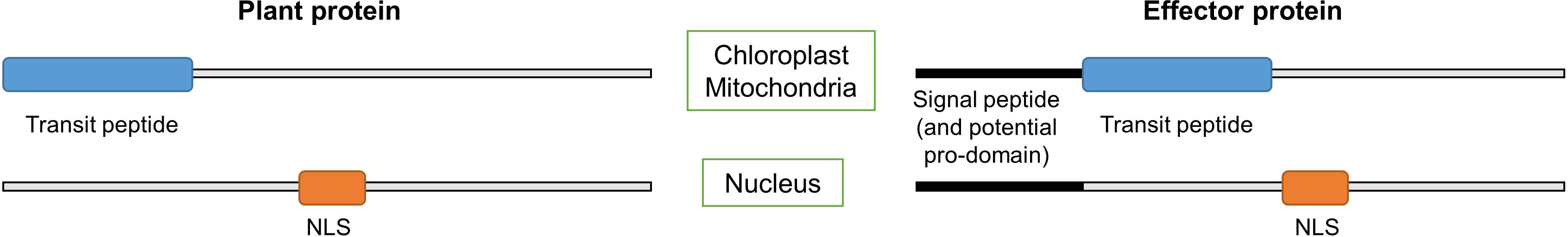
LOCALIZER is a method for predicting the subcellular localization of both plant proteins and pathogen effectors in the plant cell. It can currently predict localization to chloroplasts and mitochondria using transit peptide prediction and to nuclei using a collection of nuclear localization signals (NLSs).
Input
For plant protein localization prediction, submit full-length sequences and select plant mode.
For effector protein localization prediction, submit full-length sequences and select effector mode.
It is recommended to use tools such as SignalP or
Phobius to predict first if a protein is likely to be secreted
and to obtain the mature sequences. Alternatively, provide full sequences and let LOCALIZER delete the first 20 aas as the
signal peptide region.
Do not submit short sequence fragments to LOCALIZER, it expects the full sequence.