| Instructions | Output format | Data | Software |
LOCALIZER: subcellular localization prediction of plant and effector proteins in the plant cell
Introduction
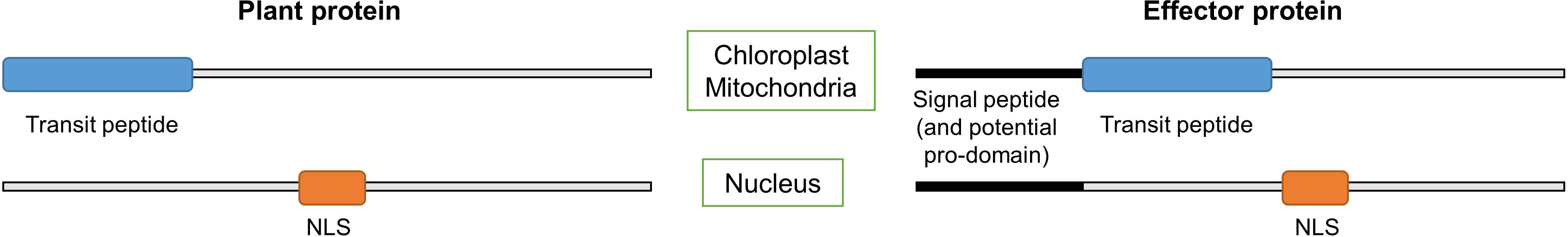
LOCALIZER is a machine learning method for subcellular localization prediction in plant cells. LOCALIZER has been trained to predict either the localization of plant proteins or the localization of eukaryotic effector proteins to chloroplasts, mitochondria or nuclei in the plant cell.
The localization to chloroplasts and mitochondria is predicted using the presence of transit peptides and the localization to nuclei is predicted using a collection of nuclear localization signals (NLSs).
Submission
*** Note: The LOCALIZER server was moved to a new server in April, which led to the use of a different BioPython version. A small number of sequences produced a prediction different to the local version (installed through GitHub) due to this. This is now fixed. ***
To run LOCALIZER, please submit your full plant or effector proteins of interest below. LOCALIZER will not use
proteins shorter than 20 aas for transit peptide prediction.
For online submission, the maximum number of protein sequences that can be submitted is 2000. If you want to run LOCALIZER on your local machine, you can download the current version here.
Citation
Contact
For comments, suggestions and reporting bugs please contact jana.sperschneider@csiro.au
